A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
The androgen receptor (AR) plays a crucial role as a transcription factor in the development of sexual characteristics and the physiological balance of tissues expressing this receptor. Despite its importance, there are several pathologies and syndromes associated with this receptor for which no definitive treatment exists. As the primary factor involved in the initiation and progression of prostate cancer, the second most common malignant disease in men in industrialized countries, the AR has been the primary therapeutic target for treating this disease for decades.
A recent study published in the journal Science Advances sheds light on the structural and functional impact of various mutations in the androgen receptor, and how these changes contribute to the development of prostate cancer. Led by Professor Eva Estébanez-Perpiñá from the Department of Biochemistry and Molecular Biomedicine of the Faculty of Biology and Institute of Biomedicine (IBUB), the study involves the participation of PhD student María Nuria Peralta Moreno and Professor Jaime Rubio Martínez from the Faculty of Chemistry and the Institute of Theoretical and Computational Chemistry (IQTC) at UB.
The theoretical group conducted a study to examine the dynamics of the androgen receptor ligand-binding domain (AR-LBD) and the potential allosteric connections between different functional regions of AR-LBD. They utilized multiple long molecular dynamics (MD) simulations to analyze both the wild-type (WT) and five different mutants located at the dimerization interface. The study started with X-ray crystallographic structures obtained for the proteins.
The MD simulations revealed that mutations in the dimerization site of AR LBD caused conformational changes in other distant pockets, indicating an allosteric coupling between dimerization and coregulator binding. Furthermore, the study quantified local conformational changes by calculating the differences in average Cα-Cα distances between spatially adjacent residue pairs, which showed that the interface mutations induced local conformational changes in AR-LBD. Interestingly, these changes occurred far from the mutated residues, suggesting an allosteric mechanism. Moreover, while some changes were observed in all mutations, specific AR regions showed markedly different local responses.
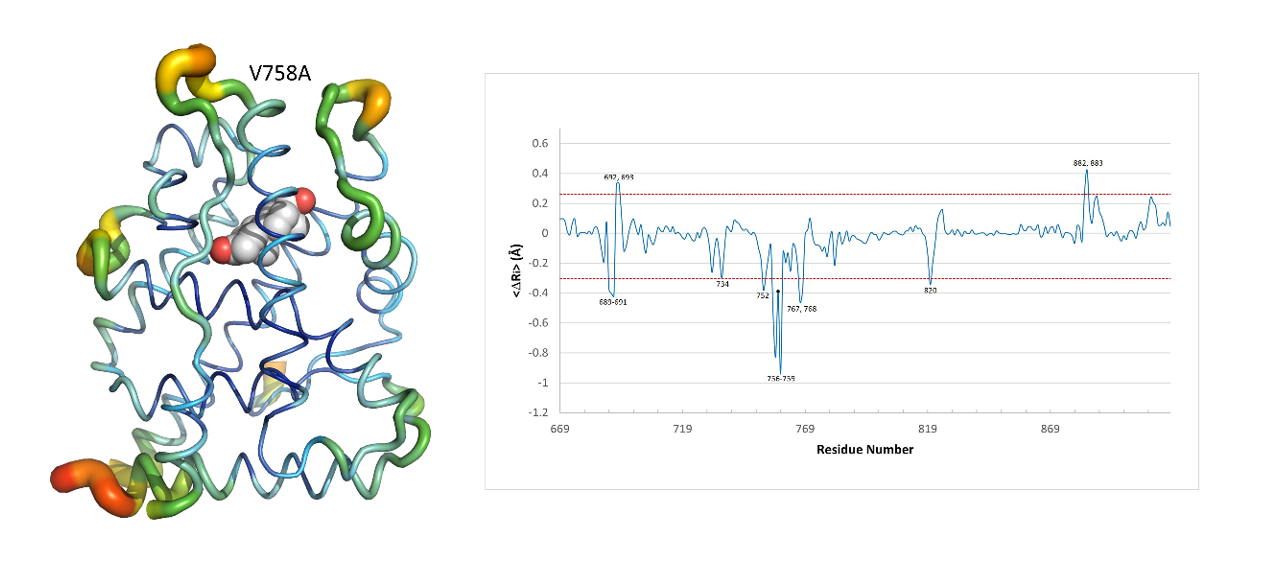
You can read the full article here: https://www.science.org/doi/10.1126/sciadv.ade2175
