Deciphering the enzymatic mechanism of sugar ring contraction in UDP-apiose biosynthesis
S. Savino, A. J. E. Borg, A. Dennig, M. Pfeiffer, F. de Giorgi, H. Weber, K. D. Dubey, C. Rovira, A. Mattevi, B. Nidetzky.
Nat. Catal., 2 (2019) 1115.
Representation of the logD of selected amino acids in the two hydrophobicity scales.
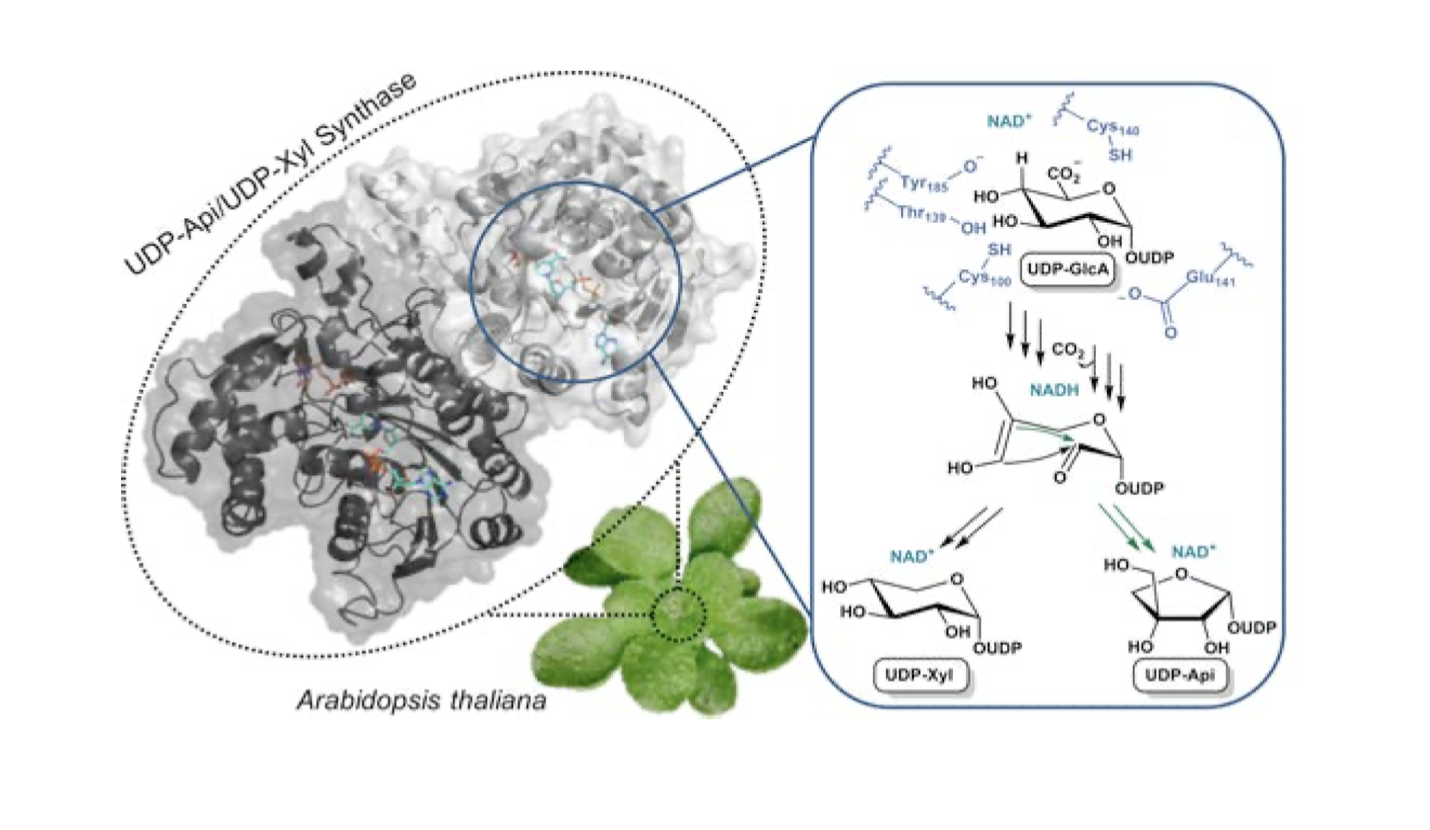
In changing weather situations, plants need to be robust and flexible at the same time. These structural properties are anchored the cell walls, which are largely built from polymers and polysaccharides. As binding agents, polysaccharides have the important task to connect long- chain polymers and to build a molecular network of tiny strands, called fibrils, which contribute to the tensile strength of the plant. One of the sugar building blocks is the branched-chain monosaccharide apiose. The mechanism responsible for apiose production of in nature was still unknown. Recently it has been discovered how apiose is produced by a single enzyme called UAXS (UDP-apiose/UDP-xylose Synthase) and the entire mechanism of this enzyme has been decoded by an interdisciplinary collaboration. The UAXS-enzyme selectively catalyzes four reaction steps, resulting in the change from a six ring sugar molecule (hexose) to a structural converted five ring sugar (pentose). By creating new organic carbon compounds, the enzyme is responsible for giving plants their strength properties.
