Development of a structure-bsed pH-dependent lipophilicity scale of amino acids from continuum solvation calculations
W. J. Zamora, J. M. Campanera, F. J. Luque
J. Phys. Chem. Letters., 10 (2019) 883.
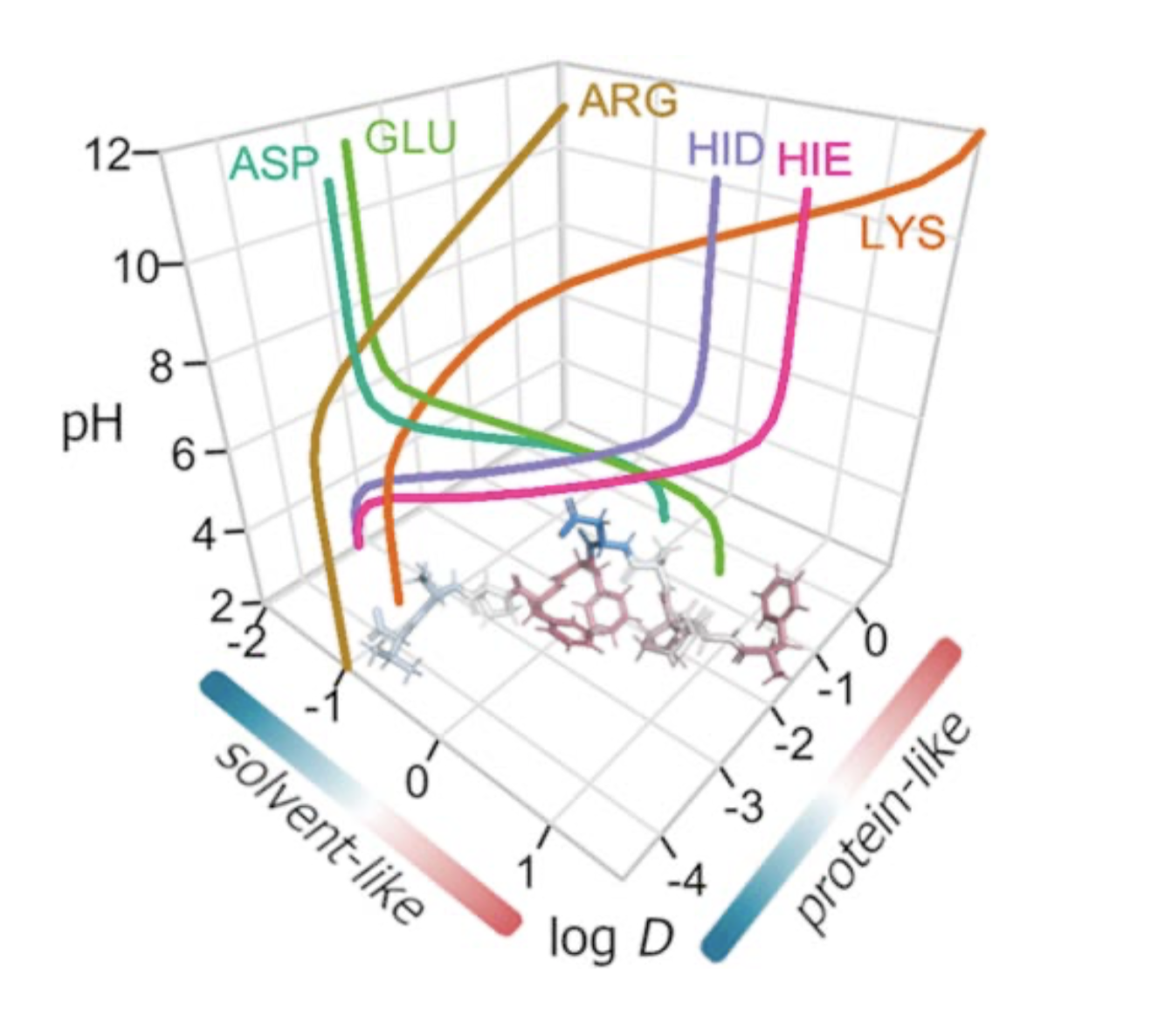
Representation of the logD of selected amino acids in the two hydrophobicity scales.
We report a versatile strategy to derive a pH-adapted scale that relies on theoretical estimates of distribution coefficients from conformational ensembles of amino acids. This is accomplished by using an accurately parametrized version of the IEFPCM/MST continuum solvation model as an effective way to describe the partitioning between n-octanol and water, in conjunction with a formalism that combines partition coefficients of neutral and ionic species of residues and the corresponding pKa values of ionizable groups. Two weighting schemes are considered to derive solvent-like and protein-like scales, which have been calibrated by comparison with other experimental scales developed in different chemical/biological environments and pH conditions as well as by examining properties such as the retention time of small peptides and the recognition of antigenic peptides. A straightforward extension to nonstandard residues is enabled by this efficient methodological strategy.
